Research
Overview
The Baker research group utilizes multidimensional separation techniques such as solid phase extractions (SPE), liquid chromatography (LC), ion mobility spectrometry (IMS), and mass spectrometry (MS) to evaluate molecules present and changing in biological and environmental systems. Research projects include the development of high-throughput analyses to study numerous samples in a short time period as well as informatics studies to evaluate and connect the complex multi-omic data with available phenotypic data.
Advanced Separations & Mass Spectrometry Methods
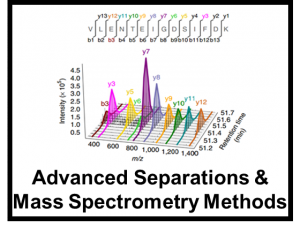
For both targeted and untargeted analyses, our lab utilizes multiple orthogonal methods of separation to resolve and identify components in a variety of complex matricies ranging from biological to environmental contexts. These methods include high performance liquid chromatography, super-critical fluid chromatography, rapid solid phase extractions, and ion mobility spectrometry coupled with high resolution mass spectrometry. As ion mobility is a gas phase separation method that occurs on the millisecond timescale, IMS is easily nested into other forms of chromatography to help separate and identify isomeric compounds in a shorter timescale. Currently, our work is mostly focused on glycomics, lipidomics, proteomics, and exposomics.
James’ work involves application of IMS-MS to the analysis of complex isomeric species in a variety of matrices. Specifically, James has applied IMS-MS to develop enhanced Newborn Screening (NBS) assays utilizing IMS-MS separations and the Agilent RapidFire solid phase extraction (SPE) platform.
Jack is working to develop a novel analytical characterization method for the analysis of Adeno-Associated Virus (AAV) capsids. Specifically, he is working in collaboration with NIST to develop an LC-IMS-MS method capable of characterizing and differentiating between various AAV serotypes, as well as their viral protein (VP) subunit components.
Environmental & Clinical Research
One of the primary focuses of our lab is the analysis of molecules in environmental and biological systems, as well as the interaction between human health and the environment . Each lab member works on implementing ongoing advancements in ion mobility spectrometry-mass spectrometry methodology and data analysis for a variety of applications to meet these goals.
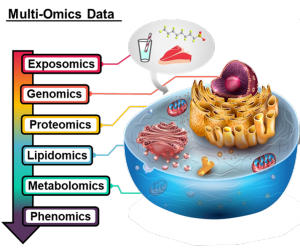
Melanie works on a number of multi-omic applications including Alzheimer’s disease, preeclampsia and COVID infection. Her main focus within her omics disease applications is lipids, which are an incredibly diverse set of molecules with a multitude of vital biological functions. However, the analytical challenges of separating out lipid isomers has until recently left lipids being drastically understudied relative to other omic fields. This has resulted in Melanie also taking up projects to assess protein-lipid interactions with collaborators all over the USA and exploring the development of informatics techniques to better annotate lipid dysregulation trends in the disease applications she is interested in.
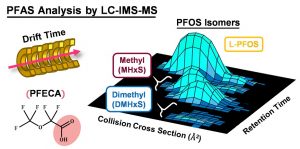
Kaylie is evaluating PFAS distribution across North Carolina over time using pine needles as passive air samplers. She is also working on investigating lipidome changes in animal models following PFAS exposure using a method which can simultaneously measure PFAS and lipids.
Anna is working on optimizing a method to extract PFAS from fish muscle tissue. Once the method is developed, she will then use LC-IMS-MS to measure PFAS in sunfish samples provided by the Belcher group.
Jessie is interested in looking at how risk for a disease is assessed based on biomarkers that are identified following a singular omics study. Her main goals are to determine how these estimates can be improved to more accurately reflect a person’s risk level, and to work on integrating data from different omics studies to create more comprehensive risk scoring schemes.
Allison is working to increase annotation confidence and metabolite coverage in microbiome studies. She is also working on using IMS-MS to increase the coverage of isomeric bile acid species and identify new bile acid conjugates in collaboration with the Theriot lab.
Rebecca is working to analyze the lipidomic effects that flame retardants have on rat brain and placenta. By analyzing the lipidomic effects, she will also determine how exposure to these flame retardants can contribute to sex-specific neurodevelopmental changes.
Data Analysis
While multidimensional experimental analyses, such as LC-IMS-MS/MS, provide specific molecular characterization and identification capabilities, annotation and interpretation of the data is severely limited by lack of software capabilities. For lipidomics data specifically, the biological interpretation of lipid data via pathway analysis requires the oversimplification of lipids to broader classes, rendering the insight from full lipid speciation irrelevant. We are working to develop comprehensive, high-throughput tools for lipid annotation, visualization and interpretation that overcome these challenges.
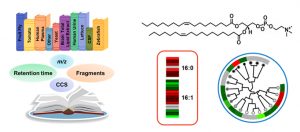
Kaylie is currently developing sample-specific spectral libraries within the free and open-source software Skyline for the rapid identification of diverse lipid species. By targeting LC, IMS, MS and MS/MS information, all lipids in the library are simultaneously annotated and visualized with a high degree of confidence. Allison is utilizing Skyline in a similar manner to rapidly characterize and confidently annotate bile acids and other metabolites of interest in microbiome studies. Rebecca is also using Skyline to characterize and annotate lipids that are found in vegetables samples such as lettuce and peanuts that were exposed to PFAS.
Melanie is working with collaborators at NCSU to understand the phenotypic implications of less studied omics, specifically how lipid variability is affected by the structural composition of lipids and how clinical information such as age, sex and diet can contribute to changes within a lipid profile. Together, these ideas are combined in our Structural Connectivity and Omic Phenotype Evaluations (SCOPE) cheminformatics toolbox for lipid data interpretation. Melanie is working to further expand the tools for our data analysis and create a more user-friendly means of integrating SCOPE for the entire lipidomics community.
Jessie is working on using machine learning to develop pipelines to select important features from large omics datasets.
Funding, Acknowledgements & Collaborations